shRNA Silencing AAV and Adenovirus
Introduction to short-hairpin RNA
Short-hairpin RNA (shRNA) is an artificial RNA molecule that derives its name from a hairpin turn in its structure. This molecule targets the transcript of the native gene for post-translational gene silencing or RNA interference (RNAi). It is an especially powerful tool to achieve specific, long-lasting gene silencing due to its low degradation rate and turnover. Due to this trait, shRNA has generated great interest for its potential application for gene therapy
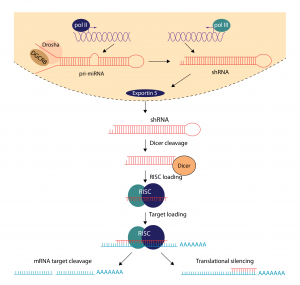
shRNA is most commonly transcribed into the nucleus by polymerase II or III. Mimicking pi-miRNA, it is then processed into pre-shRNA by Drosha. This pre-shRNA is exported from the nucleus by Exportin 5, where it is it then processed by Dicer and taken up by the RNA-induced silencing complex (RISC). At this stage, the sense strand is degraded, and the antisense strand guides RISC to a complementary mRNA sequence to silence. If the complementary sequence is perfect, the silencing complex cleaves the mRNA. If the sequence is imperfect, translation is repressed. The mechanism of silencing may differ , but the result is the same.
Viral Delivery of shRNA
Viral vectors have been the preferred the delivery method in many studies. Greater gene silencing is observed with adenovirus or AAV systems than the alternatives (direct transfection of siRNA/plasmid, or retroviral and lentiviral systems). This is due to the fact that adenovirus and AAV delivers genes into 100% of the cell population, while other systems only deliver to a fraction of that. DNA that encodes an shRNA molecule will be inserted into the adenoviral/AAV genome, in which the expression of shRNA is under the control of the U6, H1, or CMV promoter (with the option for co-expression of GFP, RFP or mCherry for easy monitoring of infection). The resulting shRNA is then rapidly converted to the desired RNAi molecule.
Single validated shRNA vs. pool shRNA
One of the main challenges of using shRNA is identifying a working shRNA sequence that can effectively knockdown the expression of the target gene. Even the best computer algorithms cannot guarantee that every shRNA candidate it generates will work inside live cells. One of the approaches for making the shRNA virus is to use a pooled shRNA solution which contains viruses expressing 3-5 shRNA to get a better mathematical chance for a good knockdown. The main benefit of using the pooled solution is cost effectiveness for screening/validating a working shRNA. However, there are two primary problems with the pooled shRNA solution:
- The knockdown efficiency from the pooled shRNA is only as good as the best shRNA in the pool, so there is no added benefit for including extra shRNAs for better silencing.
- Each additional shRNA will increase the non-specific effect of shRNA, so the pooled shRNA will amplify the non-specific effect 3-5 times more than the single shRNA.
This is why we prefer the more difficult method to make shRNA viruses: by screening shRNA candidates in cell based assays. We only use the working shRNA in the virus, so that each shRNA virus we made contains a single shRNA that has been validated in the in vitro assay.
Adenovirus Production: we will deliver a final viral stock of approximately 1010-1011 PFU. This amount is sufficient for infection of at least 200-1,000 10cm plates of most cell types. This process takes approximately 6-8 weeks to complete.
AAV Production: we will deliver a purified viral stock of approximately 1012 to 1013 genome containing particles (GC). This process takes approximately 5-6 weeks to complete.
We are able to produce shRNA adenoviruses or AAV for any gene target from any species you want to knock down.
Search our shRNA Products
Browse AAV shRNA Products
Browse Adenovirus shRNA Products
Need a Custom Virus?
We offer a wide range of viral vector services to suit your research needs.- AAV Production Service
- Adenovirus Amplification Service
- Adenovirus Construction Service
- shRNA Validation Services